Image Processing: Data based segmentation
Data-Based Segmentation: K-Means Clustering
Data-Based Segmentation involves partitioning an image into distinct regions based purely on the statistical properties of the pixel data (such as color or intensity), without using a specific geometric model (like circles or lines).
This article explores how to segment a microscopic tissue image using the **K-Means Clustering** algorithm in the L*a*b* color space.
Theoretical Background
K-Means Clustering
K-Means is an unsupervised machine learning algorithm used to partition data into distinct clusters.
- Initialization: Select random points as initial cluster centroids.
- Assignment: Assign every pixel in the image to the nearest centroid based on a distance metric (usually Euclidean distance).
- Update: Recalculate the centroids by taking the mean of all pixels assigned to each cluster.
- Repeat: Iterate steps 2 and 3 until the centroids stabilize (convergence).
Color Space Transformation
Standard RGB images mix color and lighting information, which can confuse segmentation algorithms.
- RGB (Red, Green, Blue): Not ideal for segmentation because color distance in RGB does not match human perception.
- L*a*b* (Lightness, a*, b*): A perceptually uniform color space.
- L*: Lightness (0 = Black, 100 = White).
- a*: Green to Red component.
- b*: Blue to Yellow component.
By ignoring the L* channel and clustering only on a* and b*, we segment based purely on color, making the algorithm robust to lighting variations.
MATLAB Implementation
The following MATLAB script demonstrates loading an image, converting color spaces, applying K-Means, and visualizing the result.
Code Logic
% Data-Based Segmentation
% Algorithm: K-Means Clustering (imsegkmeans)
clc; clear; close all;
% 1. Load the Image
% 'hestain.png' is a standard MATLAB demo image of H&E stained tissue
I = imread('hestain.png');
% 2. Convert to L*a*b* (Feature Extraction)
% We transform RGB to L*a*b* to isolate color information.
lab_I = rgb2lab(I);
% Extract the a* and b* channels.
% These contain the color info (Red/Green/Blue/Yellow balance).
ab = lab_I(:,:,2:3);
% Convert to single precision for the K-Means algorithm calculation
ab = im2single(ab);
% 3. Run K-Means
% We choose k=3 because H&E stained images typically contain:
% 1. Purple (Nuclei)
% 2. Pink (Cytoplasm/Tissue)
% 3. White (Background)
k = 3;
pixel_labels = imsegkmeans(ab, k, 'NumAttempts', 3);
% 4. Visualization
figure('Name', 'Task 4: Segmentation Results', ...
'NumberTitle', 'off', ...
'Position', [100, 100, 1000, 500]);
% Show Original
subplot(1, 2, 1);
imshow(I);
title('Original Image');
% Show Segmentation Label Matrix
subplot(1, 2, 2);
imagesc(pixel_labels);
axis image;
colormap(gca, jet); % 'jet' gives distinct colors for 1, 2, and 3
colorbar;
title(['K-Means Clusters (k=', num2str(k), ')']);
Analysis of Results
The result, stored in pixel_labels, is a matrix of the same size as the image where every value is 1, 2, or 3.
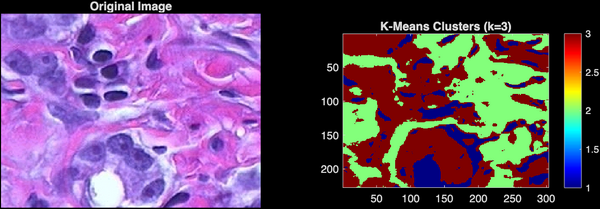
Interpretation
- Cluster 1: Typically corresponds to the white background (low a*, low b*).
- Cluster 2: Typically corresponds to the pink connective tissue (medium a*).
- Cluster 3: Typically corresponds to the purple nuclei (high a*, low b*).
Note: The specific label numbers (1, 2, 3) depend on random initialization and may swap between runs.
Why use 'NumAttempts'?
K-Means can get stuck in local minima depending on where the initial centroids are placed. By setting 'NumAttempts', 3, MATLAB runs the algorithm 3 times with different random starting points and returns the result with the lowest total error summation.
Key MATLAB Functions
rgb2lab: Converts images from RGB to CIE L*a*b* color space.imsegkmeans: A specialized function for image segmentation using K-Means (optimized for image data structures).imagesc: Displays data as an image using the full range of the current colormap (useful for viewing label matrices).